Not just curiosities - Ubiquitin-independent protein degradation
The Ubiquitin-Proteasome (UPS) system is the major route by which the cell degrades unwanted proteins. Ubiquitin is a small protein that tags proteins for degradation via a protein complex called proteasome. The addition of ubiquitin to the protein targeted for degradation occurs through a cascade of three enzymes - E1 which activates the ubiquitin protein, E2 which transfers the ubiquitin to the target protein that is recruited by E3. The E3 enzyme provides specificity by recognizing a short motif in the protein to be degraded called a degron.
So far reports have indicated a small number of proteins that are able to undergo ubiquitin-independent proteasomal degradation (UbInPD). For many of these proteins, the evidence supporting ubiquitin-independent degradation was obtained in vitro (in a test tube), and hence, the biological relevance of this degradation remains unclear. Over the years, efforts have been made to characterize the molecular mechanisms by which substrates undergoing UbInPD are targeted to the proteasome. However, given the relatively small number of substrates identified, conclusions have been drawn on a case-by-case basis.
In groundbreaking work, published in the prestigious journal "Molecular Cell", a group of researchers from Israel and the world led by Dr. Itay Koren from the Mina and Everard Goodman Faculty of Life Sciences at Bar-Ilan University showed that the UblnPD pathway is more prevalent than currently appreciated and succeeded to identify specific protein sequences (motifs) that promote it.
In their previous studies, Dr. Itay Koren and his colleagues developed and used a platform for degron identification named GPS-peptidome to investigate the specificity of degradation in the UPS. GPS-peptidome is a method that measures protein stability combined with a synthetic representation of the human peptidome (a complete set of peptides, a short chain of amino acids, encoded by a genome). Using this platform, researchers identified a suite of degrons located at the carboxyl-termini of human proteins. While the E3 enzymes family regulated the majority of degrons that were identified, few, with amino acid alanine or valine at the degron's end, were not.
To characterize the degradation mode of proteins bearing these motifs, the researchers selected three representative proteins - CPS1, SREBF2, and OR4C13 and fused their 23 amino acid long (C23mer) degrons bearing the special motif to a green fluorescent protein (GFP). Next, cells expressing the GFP-fusions were treated with either MLN7243, which inhibits E1 enzymes, thus eliminating ubiquitinylation, or with proteasome inhibitor bortezomib, and the stability of the fussed GFPs was monitored. The stability of the GFP-C23mer fusions increased in response to proteasome inhibitor, but remained unchanged or decreased following E1 inhibitor treatment. Similar results were obtained with GFP fusion with the C-terminus of a protein known to undergo ubiquitin-independent degradation. GFP that was fused to C-terminus from a protein that undergoes ubiquitin-dependent degradation was stabilized with both inhibitors.
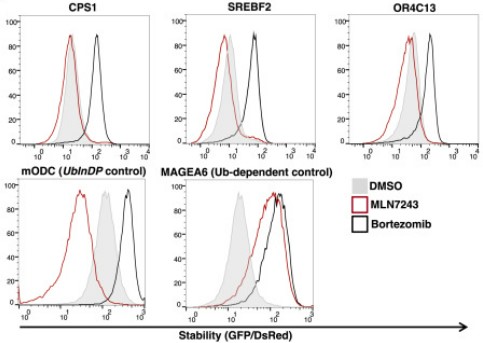 In the figure - cells expressing a GFP protein fused to the last 23 amino acids of the proteins CPS1, SREBF2 or OR4C13 were treated with MLN7243, which inhibits the E1 enzymes, or with the proteasome inhibitor bortezomib, or with a control treatment (DMSO). As a control, GFP protein was fussed to the carboxyl terminus of the mODC protein known to undergo UblnDP or the MAGEA6 protein known to undergo ubiquitin-dependent degradation. The GFP/dsRed ratio represents the stability of the indicated GFP-fusion proteins in the presence of the indicated inhibitors - a sharp peak to the right side indicates stabilization.
In the figure - cells expressing a GFP protein fused to the last 23 amino acids of the proteins CPS1, SREBF2 or OR4C13 were treated with MLN7243, which inhibits the E1 enzymes, or with the proteasome inhibitor bortezomib, or with a control treatment (DMSO). As a control, GFP protein was fussed to the carboxyl terminus of the mODC protein known to undergo UblnDP or the MAGEA6 protein known to undergo ubiquitin-dependent degradation. The GFP/dsRed ratio represents the stability of the indicated GFP-fusion proteins in the presence of the indicated inhibitors - a sharp peak to the right side indicates stabilization.
To systematically search for additional C-degrons promoting UbInPD, researchers applied the method they developed on cells treated with either E1 inhibitor or proteasome inhibitor, or left untreated, and identified 1,829 ubiquitin-independent and 2,121 ubiquitin-dependent degrons. Researchers validated the results by examining the stability of 13 degrons, identified as ubiquitin-independent, in the presence of inhibitors. Findings indicated that all of the degrons were stabilized in proteasome-inhibitor-treated cells but degraded in E1 inhibitor-treated cells. The data suggests that when E1 is inhibited, the proteasome's capacity to degrade proteins from the ubiquitin-independent pathway increases.
Sequence analysis of ubiquitin-dependent C23mer substrates revealed alanine, cysteine, and valine enrichment in their extreme end. To experimentally validate the importance of those motifs in mediating UbInPD, researchers mutated approximately 100 UbInPD C23mer by either replacing or deleting those alanine, cysteine, and valine amino acids. The mutagenesis revealed that indeed those amino acids are involved in peptide degradation - mutation or deletions of them increased peptide stability in the presence of inhibitors. Interestingly, repositioning of the terminal motif by adding additional amino acids after the motifs leads to peptide stabilization, indicating the importance of the motif being exposed at the C terminus to promote UbInPD.
In in vitro experiments, both 20S, and 26S proteasomes have been shown to mediate the degradation of UbInPD substrates. In order to understand which of the proteasomes promote UbInPD in cells (in vivo), researchers depleted PSMD1, a proteasome 26S subunit that is required for the proteasome 26S activity but not for the 20S proteasome activity. In the PSMD1-depleted cells, the UbInPD substrates (the GFP-C23mer fusions) were significantly stabilized, indicating that in vivo UbInPD depends on 26S and not 20S proteasome.
Up to this point, researchers screened and investigated peptide substrates (23mers) and their GFP-fusions. To elucidate the biological significance and molecular mechanism of UbInPD, the researchers were interested in identifying full-length protein substrates. They therefore screened a library of approximately 15,000 constructs encoding full-length human proteins in proteasome-inhibitor-treated or E1 inhibitor-treated cells. This approach identified 69 full-length proteins as candidate UbInPD substrates, with few of them previously established as UbInPD substrates. The relatively small number of substrates compared with the peptidome screen suggests that in the context of full-length proteins, only a specific subset of proteins are degraded in a ubiquitin-independent manner.
Next, a few of the identified proteins were validated individually as either GFP fusions or independent proteins in proteasome or E1-inhibited cells. Researchers found that all the validated proteins stabilized in proteasome-inhibited cells, indicating that their degradations depend on the proteasome. Following treatment with E1 inhibitor part of the protein showed no increase or reduced protein level, indicating to be UbInPD substrates. This finding suggests that not only peptides but also full-length proteins can be targeted for ubiquitin-independent degradation.
To examine whether the C terminus does play a role in the full-length proteins that were identified as UbInPD substrates, researchers either deleted or added extra amino acids to proteins' C terminus and assayed the impact on the proteins' stability. The mutated proteins exhibited stabilization upon treatment with both proteasome and E1 inhibitors, suggesting that those proteins can be degraded in both ubiquitin-independent and ubiquitin-dependent manner, and, thus, when one mode of degradation is attenuated, these substrates can be degraded in the other pathway.
Finally, the researchers sought to understand the mechanism by which UbInPD substrates are directed to the proteasome. Researchers hypothesized that Ubiquitin-like (UBL) proteins, that bridge substrates to the proteasome by associating with the proteasomal ubiquitin receptors, might be involved in targeting UbInPD substrates as well. By deleting different UBL proteins, researchers found that only after triple deletion of the Ubiquilin family proteins (UBQLN1, UBQLN2, and UBQLN4) in combination with E1 inhibition, were UbInPD substrates stabilized. In addition, researchers demonstrated physical interaction between UBQLN1 and the UbInPD protein substrate and mapped the region in the UBQLN1 that promotes the UbInPD.
Overall, the study represents a systematic approach to investigating UbInPD, showing that UbInPD is more prevalent than currently appreciated. More studies are needed in order to decipher this important and complex mechanism.
This work was published in "ScienceDirect".
Last Updated Date : 12/11/2024