Prof. Yaron Shav-Tal gets up close and personal with living cells
Prof. Yaron Shav-Tal, head of the Dangoor Center for Personalized Medicine at Bar-Ilan University, closely observes cell activity and the life cycle of RNA within it. Interestingly, this has implications for cancer research.
What can you discover by observing a live cell up close? Prof. Yaron Shav-Tal, Director of the Dangoor Center for Personalized Medicine and Vice President for Research at Bar-Ilan University, researches the field of gene expression. His team monitors RNA activity and other molecules within the living cell in real-time, using sensitive microscopic technologies and fluorescent molecule labels.
Prof. Shav-Tal, why is it necessary to label molecules? What can be seen when looking at a cell under a microscope without staining?
"The cells themselves are transparent. Apart from red blood cells, which are truly red, all others are colorless, so if we place a cell under a microscope, we won't see much. We'll see how parts of the cell move, but we won't get much information about the processes happening inside—and there's a lot happening: construction, breakdown, movement. If you want to observe dead cells, you can pour a substance on them that kills and fixes them, and then stain the molecules. However, with this method, you lose the entire dimension of real-time motion."
And you want to see the living cells in motion.
"Yes. Because when I look at a dead cell, I can see its various parts, but I can't learn anything about its dynamics and kinetics. That’s why researchers focus on different methods for labeling molecules in the cell, and these methods have revolutionized science. There are three major milestones in the development of protein labeling in cells, and Nobel Prizes were awarded for each of them."
 Green Fluorescent Protein (GFP)
Green Fluorescent Protein (GFP)
Jellyfish in the service of personalized medicine
How were the methods for labeling cell components discovered?
"The initial discovery, which revolutionized biology, was the identification of a molecule that emits green fluorescent light, called GFP (Green Fluorescent Protein), derived from a protein found in jellyfish. This discovery earned the Nobel Prize in 2008. In the second stage, it was discovered that the jellyfish genome is composed of the same letters as those found in the genes of every living organism. This meant it was possible to attach the fluorescent molecule to the genes of worms, mice, and humans, creating a hybrid gene that continues to perform its functions while carrying the fluorescent tag, enabling researchers to track its movement within the cell."
"Thanks to this invention, any researcher in the world can attach this gene to the molecule they are studying and observe it under the microscope. If I want to track a cancerous tumor in an animal like a mouse, I can stain the cancerous cells and see whether the tumors grow or shrink following chemotherapy treatment. Some even turned entire animals, like cats or rabbits, into glowing green creatures.
 A fluorescent green axolotl fish
A fluorescent green axolotl fish
Sounds terrible. Was that done to humans as well?
"Theoretically, it’s possible to do it to humans, but it’s not done."
And the third stage?
"In the third stage, mutations were made in the protein, creating different colors. From the green GFP, they developed yellow YFP and red RFP. Now, it’s possible to use the different colors and attach them to different molecules so that each molecule has a distinct color, allowing researchers to track them simultaneously."
Are these also the methods you use to observe RNA?
"Not exactly. In fact, these methods by themselves do not allow us to observe RNA because, like DNA, RNA is composed of nucleic acids and not amino acids. Therefore, it’s impossible to embed the dyes, which are also made of amino acids, onto them. For this reason, for many years, there was no way to track RNA and DNA; they didn’t know how to stain them. The method we use was developed at the Einstein School of Medicine in New York, where I conducted my postdoctoral research. It takes advantage of the fact that as RNA moves around the nucleus, it collects many proteins. Some of these proteins can be labeled, creating a tail of dozens of fluorescent proteins. Essentially, under the microscope, we don’t see the entire RNA strand as a thread but only its tail, which is labeled."
What does the cell's RNA look like under the microscope?
"Under the microscope, you don’t see the transparent parts; they are darkened, and only the illuminated parts are visible. For example, if the microscope is set to blue light, which is meant to illuminate the green fluorescence, you’ll only see the molecules labeled in green, with complete darkness surrounding them. The RNA itself appears as small dots moving within the living cell."
The short and difficult life of messenger RNA
What do you discover when tracking RNA movement in the cell?
"The purpose of messenger RNA - mRNA, is to transfer the genetic information found in the cell nucleus to the cytoplasm, where all the vital activities of the cell occur, based on instructions from the genetic material. The separation between the nucleus and the body of the cell is unique to humans and other animals; bacteria, for example, don’t have a nucleus. In bacteria, all parts of the cell coexist without partitions, and the genetic material in the DNA immediately goes into action. But in humans, the DNA is confined to the nucleus and cannot move. The RNA takes on the role of transferring the information to the cytoplasm. It copies the information from the DNA and crosses the 'barrier' to the cytoplasm, where a ribosome attaches to it, reads it, and creates a protein that subsequently operates within the cell."
How does RNA make the journey out of the nucleus?
"RNA has a life cycle, somewhat like humans, except its typical lifespan is only a few minutes. Even before it exits, while it is still 'young,' there is a system in the nucleus that checks it to ensure its transcription is correct. If there is an error or mutation, it will not be allowed to leave."
What will happen to it?
"It will be blocked, mature inside the nucleus, and die there. Enzymes known as 'predators' roam the cell, spotting RNA and destroying it. Cells don’t want RNA to have a long life, because they need to respond in real-time to what’s happening around them. On the other hand, the cell doesn’t want RNA to be eliminated too quickly, so the RNA is covered with a layer of hundreds of proteins that protect it from these predators. At this stage, the RNA no longer looks like a strand but like a particle, and that’s how it moves around the nucleus. It’s this protective layer that allows us to see it."
How does RNA exit the nucleus?
"RNA has to find its own way out of the nucleus. The nuclear envelope has many openings called 'nuclear pores,' which serve as exit doors from the nucleus. However, the nucleus is packed with DNA and chromosomes that occupy most of the space, and some of it is so densely packed that RNA cannot pass through, or it must squeeze between numerous strands. We studied this years ago and found that RNA’s movement within the nucleus is non-directed. Although it needs to move from the nucleus to the cytoplasm, there is no guidance, and its progress is through diffusion – which is completely random. This means it takes time for it to locate the openings. RNA is also relatively long and quite heavy; it’s like a small elephant wandering in the nucleus, and it takes time for it to find its way."
Did you see it squeezing through the pore?
"Yes, that is what made us unique: we were the first in the world to see RNA exiting through the pore. No one had managed to observe this before us. Only afterward did others come and conduct similar studies. We discovered that after RNA slowly wanders through the nucleus, the moment it passes through the pore, it looks as if someone kicks it out at incredible speed. Just after presenting its 'entry ticket' to the gatekeeper, showing that it is mature and intact, it shoots out in fractions of a second."
Why couldn’t others see it?
"You can’t see it without observing the cell in real-time. With conventional methods, researchers study RNA in cells that have long been dead, but the moment the cell dies freezes things in a specific state, without revealing what happened before or after. Therefore, experiments on dead cells show only a single point in time and not the cell over time. Typically, researchers take millions of cells, chop them up in a test tube, and get an average picture of the cell population. We approach things differently: I want to know what happens in a single specific cell."
The cell's horror movie
Let’s continue with RNA – what happens to it in the cytoplasm?
"As we mentioned, it encounters the ribosome there, which reads it and produces proteins based on it. But RNA’s time is short: the predator enzymes are also present in the cytoplasm, and after it has been translated several times, the enzymes will eventually target and destroy it. This is because, as the process progresses, the RNA loses some of the proteins that protect it, and its defense weakens."
Does it also wander slowly in the cytoplasm?
"No. In some cells, we see that significant attention is given to transporting RNA from one place to another. For example, a giraffe has a very long cell that starts in its brain and extends all the way down to the tip of its leg. If we relied on diffusion to move molecules within the cytoplasm of that cell, the process could take months. So, in the cytoplasm, there is a transport system, like 'shuttles,' that move the particles from place to place."
What are the implications of your research, beyond understanding the basic processes of the cell?
"Alongside studying the normal processes within the cell, we also aim to investigate abnormal processes. For instance, RNA that reaches the cytoplasm is typically translated and dies within a short time. However, under certain stress conditions, some RNA enters 'granules' – large storage structures within the cytoplasm. We discovered that the cell hides RNA there to protect it from changes occurring during stress, as the cell wants to quickly resume translating RNA once the stress subsides, so it needs readily available RNA."
What do you mean by "stress conditions"?
"For example, chemotherapy is a stress condition for the cell. This led to the hypothesis that one reason some cancer cells survive chemotherapy is that they hide some of their RNA molecules within granules. Researchers are trying to disrupt the cell's ability to produce granules for RNA, for instance, by administering substances that prevent granule formation or prevent granules from breaking down after the stress. This would create a 'pile of waste' within the cancer cell, causing it to die."
 Prof. Yaron Shav-Tal
Prof. Yaron Shav-Tal
You’re practically screenwriters in the cell’s movie.
"Yes. Let me give you another example of such a storyline. We have a student in the lab working with herpes viruses, studying what happens to a cell when it gets infected. After the virus infects the cells, you can see the nucleus starting to produce the virus’s DNA, and all its contents and structure change. We’re investigating what happens in the nucleus a few hours after the virus infects the cell, and it’s absolute chaos. It changes the entire cell and ultimately kills it. This process takes a few hours, and it really looks like a horror movie."
The cost of microscope
Prof. Shav-Tal completed his PhD at the Weizmann Institute and conducted his postdoctoral research at the Einstein School of Medicine in New York. Since 2005, he has been part of the The Mina & Everard Goodman Faculty of Life Sciences at Bar-Ilan University. He has published over 100 articles and received numerous awards and research grants, including prestigious grants from the European Research Council (ERC) and the National Institutes of Health (NIH) in the United States. For about a decade, he served as Vice Dean of the Faculty of Life Sciences. Two years ago, he was appointed Dean, and recently he became the University's Vice President for Research and the Head of the Dangoor Center for Personalized Medicine.
Why did you choose to establish your lab at Bar-Ilan?
"As is customary, I toured the universities and felt that Bar-Ilan was experiencing significant momentum, both in building infrastructure and in its plan to recruit many young researchers over a decade. I knew it was a place where growth was possible, and the atmosphere was good. I can say that I am happy with my choice. Today, science requires a great deal of funding – for purchasing equipment, materials, and more. Microscopes like the ones we use can cost at least $300,000, and they need to be replaced every few years. The university supported me in my research endeavors."
As the new head of the Dangoor Center, what is your vision?
"In addition to recruiting new researchers to work at the forefront of personalized medicine, we plan to expand collaborations with the Sheba Medical Center and issue calls for proposals to foster partnerships between researchers and physicians from both institutions, integrating science and clinical practice. We aim to extend these collaborations into additional fields of knowledge beyond life sciences that intersect with various areas of medicine."
Finally, looking ten years ahead, what would you like to see advancing in your field?
"I can't predict, but I would be delighted if, in the future, I could use a microscope to observe cells within the organism itself – meaning to track cells in their natural tissue rather than just in the lab."
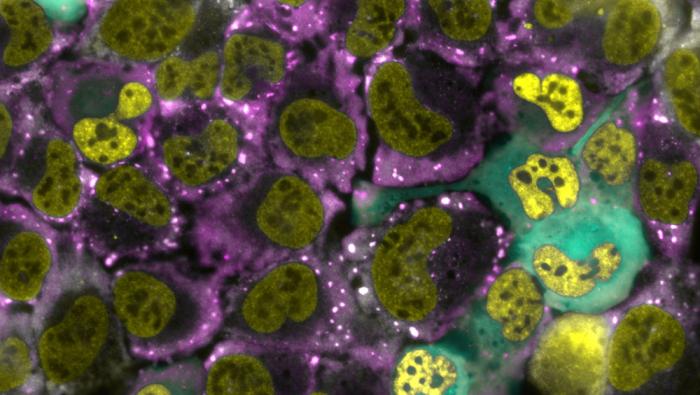
Main photo: Bone cancer cells from a human. In yellow: RNA in the cell nucleus. In purple: cytoplasm. In blue: cells in which the researchers caused RNA to become trapped in the nucleus. The white dots are the granules formed under stress conditions. Photo credit: PhD student Muhammad Atrash
Last Updated Date : 05/01/2026