Learning about Sex Determination on a Dish - No Longer Science Fiction
During embryonic development the identity of the bipotential gonad, and its potential to develop into a male (testis) or a female (ovary), is determined by a cascade of signals that antagonize each other. Errors in this process result in disorders of sex development (DSDs), characterized by the discordance between chromosomal, gonadal (testis or ovary), and anatomical sex. Currently, our knowledge and understanding of sex-determination stem from mice embryos and genetic analysis of humans with errors of sex determination. However, the mouse model is limited due to insufficient evolutionary conservation in sex determination, while the human cell models that have been proposed do not faithfully mimic the natural cells. Thus, the absence of an appropriate and accessible in vitro research system creates a major obstacle in understanding the mechanisms of sex-determination and DSDs.
An international team of researchers led by Dr. Nitzan Gonen, from the Mina and Everard Goodman Faculty of Life Sciences and the Nanotechnology Institute at Bar-Ilan University, has developed a powerful model that enables the study of testis determination and differentiation in mouse and human, as well as the study of cellular and molecular effects of genes associated with DSDs.
First, the researchers worked with mouse stem cells, developing a protocol that mimics the known pathway of gonad development, by using well-defined media and adding growth factors and small molecules in a highly timed manner. The researchers differentiated stem cells (cells that have the potential to become almost any cell in the body) into gonadal progenitor cells (cells that will differentiate into testicular or ovarian cells). Throughout the differentiation process, the researchers examined the expression of specific markers of each stage of the differentiation, from stem cells to gonadal progenitor cells, both at the RNA transcripts and proteins level. In addition, the researchers compared the gene expression of the gonadal progenitor cells generated to the gene expression dataset on mouse embryos' gonads and found that the cells they created are similar to gonadal progenitors of the male (XY) and female (XX) gonads of 11.5 days embryo.
Next, the researchers went one step further, differentiating the gonadal progenitor cells they generated into Sertoli-like cells, testicular cells that are essential for testis formation and the sperm production process. In order to induce differentiation into Sertoli cells, the genes Nr5a1 and Dmrt1, which are known to be important for differentiation into Sertoli cells, were inserted into the progenitor cells. The mouse stem cells the researchers chose to work with contain a fluorescent protein with a cyan color wavelength that is expressed when the cells become Sertoli cells. As a result of the expression of the Nr5a1 and Dmrt1 genes, the researchers obtained cyan fluorescent-positive cells. Moreover, these cells formed tubule-like structures, reminiscent of the structure of testis cords, normally present in the testis.
Then, the researchers applied the developed Sertoli-like cells differentiation protocol on human stem cells and tested whether this protocol would enable them to generate a model for studying male-to-female sex reversal DSD pathology. For this purpose, the researchers worked with stem cells generated from a healthy male (XY), a healthy female (XX), and a person who is genetically male but as a result of a mutation in the Nr5a1 gene suffers from DSD (46,XY DSD) and appears as female. The physical appearance (phenotype) of mutations in the Nr5a1 gene is variable and ranges from a complete absence of testis to normal-shaped but infertile testis. During the process of differentiation into Sertoli cells, cells exhibit changes in shape and gene expression, but at the end of the differentiation, only healthy male (46,XY) cells showed rearrangement and aggregation in the dish. In addition, only the 46,XY cells formed the tubular structures characteristic of Sertoli cells when seeded atop a soft substrate.
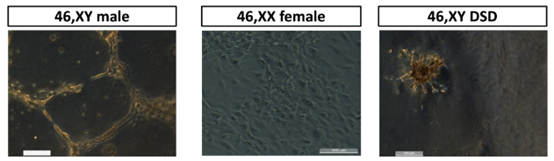 In the figure - human stem cells were differentiated into Sertoli cells and seeded on a soft substrate. On the left - differentiated cells from a healthy male (46,XY), in the middle - differentiated cells from a healthy female (46,XX) and on the right, differentiated cells from a male DSD patient with a mutation in the Nr5a1 gene (46,XY DSD). Only healthy male cells formed the tubular structures characteristic of Sertoli cells.
In the figure - human stem cells were differentiated into Sertoli cells and seeded on a soft substrate. On the left - differentiated cells from a healthy male (46,XY), in the middle - differentiated cells from a healthy female (46,XX) and on the right, differentiated cells from a male DSD patient with a mutation in the Nr5a1 gene (46,XY DSD). Only healthy male cells formed the tubular structures characteristic of Sertoli cells.
To evaluate the capacity of the generated cells to migrate, aggregate, and form 3D structures, researchers designed a microfluidic device with three channels: one central for the soft substrate loading and two lateral channels for media and cell loading. The differentiated human Sertoli-like cells were seeded into the microfluidic device and the cells were imaged for 72 hours. The live imaging movies showed that all the cells survived and proliferated. The healthy and DSD male cells (46,XY and 46,XY DSD) migrated and invaded the central channel filled with a soft substrate, whereas the female cells (46,XX) cells didn't. However, only the 46,XY cells migrated in a coordinated manner leading to recognizable tubular structures, while the 46,XY DSD cells were less coordinated and didn't form tubular structures.
Finally, the researchers wanted to test whether it was possible to obtain normal Sertoli cells from DSD male-derived stem cells (46,XY DSD) that were corrected using CRISPR, a genome editing technology. The researchers differentiated the edited cells using a developed protocol and tested the expression of RNA transcripts and protein markers of male gonads in these cells. The expression of the markers in the corrected and differentiated cells resembles the expression in normal and differentiated male cells (46,XY). Moreover, when seeded on a soft substrate the edited cells formed the same tubular structures as cells from a healthy male.
The model developed by Dr. Gonen and her colleagues is an important tool in understanding the molecular processes of testis differentiation and DSDs and potentially in understanding processes related to fertility issues.
This work was published in the prestigious journal "Science Advances".
Last Updated Date : 12/11/2024